Publications
Beeby, Morgan, Josie L. Ferreira, Patrick Tripp, Sonja-Verena Albers, and David R. Mitchell.
Propulsive Nanomachines: The Convergent Evolution of Archaella, Flagella, and Cilia.
2020. FEMS Microbiology Reviews.
View
Tadija Kekic & Ivan Barišić.
In Silico Modelling of DNA Nanostructures.
2020. Computational and Structural Biotechnology Journal.
View
Yasaman Ahmadi, Ashley L Nord, Amanda J Wilson, Christiane Hütter, Fabian Schroeder, Morgan Beeby, Ivan Barišić.
The Brownian and Flow-Driven Rotational Dynamics of a Multicomponent DNA Origami-Based Rotor.
2020. Small, e2001855.
View
Tsai C.-L., Tripp P, Sivabalasarma S, Zhang C, Rodriguez-Franco M, Wipfler RL, Chaudhury P, Banerjee A, Beeby M, Whitaker RJ, Tainer JA, Albers SV.
The structure of the periplasmic FlaG-FlaF complex and its essential role for archaellar swimming motility.
2020. Nature Microbiol, 5(1):216–225.
View
Tadija Kekic, Yasaman Ahmadi, Ivan Barišić.
An Enzymatic Active Site Embedded in a DNA Nanostructure.
2019. BioRxiv.
View
Elisa de Llano, Haichao Miao, Yasaman Ahmadi, Amanda J. Wilson, Morgan Beeby, Ivan Viola, Ivan Barišić.
Adenita: Interactive 3D modeling and visualization of DNA Nanostructures.
2019. BioRxiv.
View
Yasaman Ahmadi, Ivan Barišić.
Gene-therapy Inspired Polycation Coating for Protection of DNA Origami Nanostructures.
2019. J. Vis. Exp. (143)
View
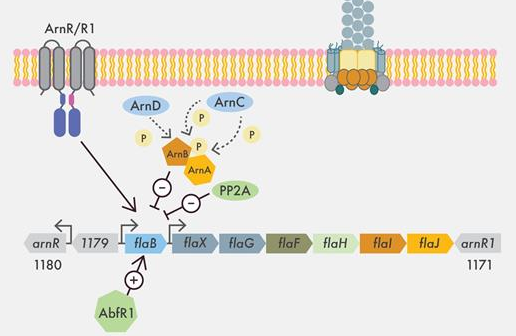
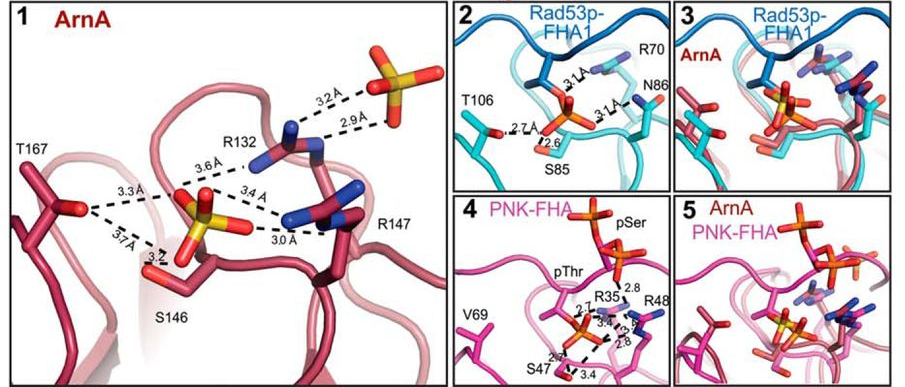
Hoffmann L, Anders K, Bischof LF, Ye X, Reimann J, Khadouma S, Pham TK, van der Does C, Wright PC, Essen LO, Albers SV.
Structure and interactions of the archael motility repression module ArnA-ArnB that modulates archaellum gene expression in Sulfolobus acidocaldarius.
2019. J Biol Chem, 294:7460-7471
View
Li Z, Kinosita Y, Rodriguez-Franco M, Nußbaum P, Braun F, Delpech F, Quax TEF, Albers SV.
Positioning of the motility machinery in halophilic archaea.
2019. MBio, 10(3)
View
Ahmadi, Y., De Llano, E., Barišić, I.
(Poly)cation-induced protection of conventional and wireframe DNA origami nanostructures.
2018. Nanoscale 10, 7494–7504.
View
Chaudhury, P., Tripp, P., Albers, S.-V.
Expression, Purification, and Assembly of Archaellum Subcomplexes of Sulfolobus acidocaldarius.
2018. Marsh, J.A. (Ed.), Protein Complex Assembly. Springer New York, New York, NY, pp. 307–314.
View
Miao, Haichao, De Llano, E., Sorger, J., Ahmadi, Y., Kekic, T., Isenberg, T., Groller, M.E., Barisic, I., Viola, I.
Multiscale Visualization and Scale-Adaptive Modification of DNA Nanostructures.
2018. IEEE Transactions on Visualization and Computer Graphics 24, 1014–1024.
View
Miao, H., Llano, E.D., Isenberg, T., Gröller, M.E., Barišić, I., Viola, I.
DimSUM: Dimension and Scale Unifying Map for Visual Abstraction of DNA Origami Structures.
2018. Computer Graphics Forum 37, 403–413.
View
Haurat, M.F., Figueiredo, A.S., Hoffmann, L., Li, L., Herr, K., J. Wilson, A., Beeby, M., Schaber, J., Albers, S.-V.
ArnS, a kinase involved in starvation-induced archaellum expression: Starvation-induced archaellum expression.
2017. Molecular Microbiology 103, 181–194.
View
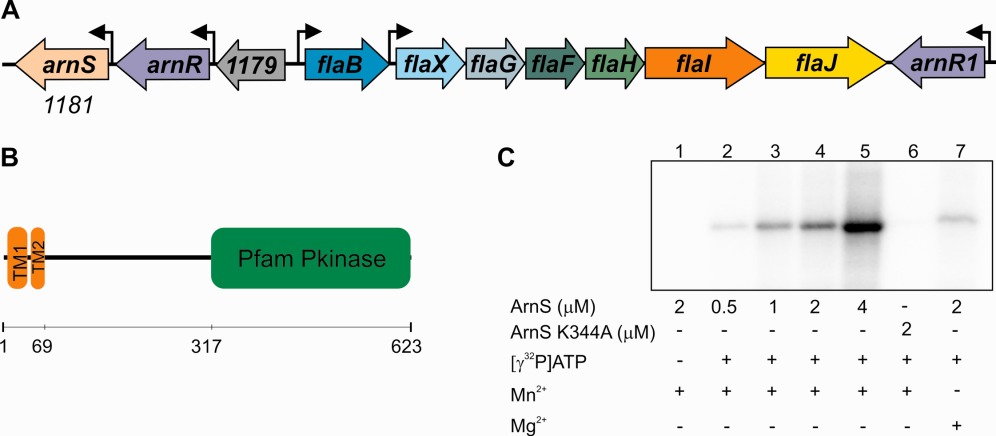
Hoffmann, L., Schummer, A., Reimann, J., Haurat, M.F., Wilson, A.J., Beeby, M., Warscheid, B., Albers, S.-V.
Expanding the archaellum regulatory network - the eukaryotic protein kinases ArnC and ArnD influence motility of Sulfolobus acidocaldarius.
2017. MicrobiologyOpen 6, e00414.
View
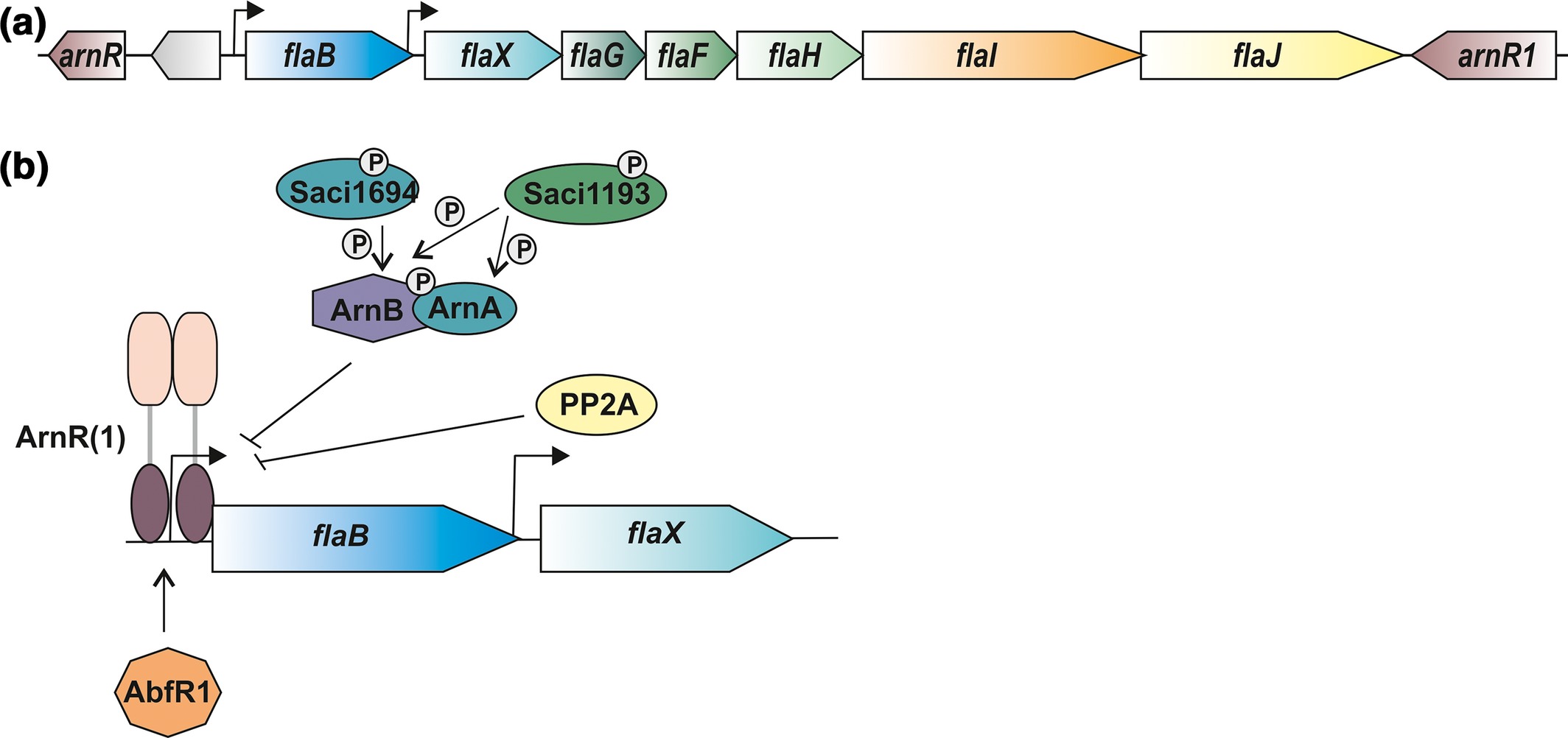
Esser, D., Hoffmann, L., Pham, T.K., Bräsen, C., Qiu, W., Wright, P.C., Albers, S.-V., Siebers, B.
Protein phosphorylation and its role in archaeal signal transduction.
2016. FEMS Microbiology Reviews 40, 625–647.
View